Making it hard for INFLUENZA A VIRUS to replicate
LOFlu Project - controlling influenza A virus liquid organelles
Sponsored by

Sponsored by

Viral outbreaks need a first line of antiviral strategies to tackle emerging viruses since the on-demand development of vaccines or antibody treatment is not always feasible. To address this problem, the Universidade Católica’s Biomedical Research Centre has been investigating the host-pathogen interactions in influenza A virus (IAV) infection.
Led by Maria João Amorim and funded by the EU, the LOFlu project places particular emphasis on viral inclusions that form during viral assembly and constitute a key step in the virus life cycle. These sites concentrate viral genetic material in specific locations in the cytosol, and are thought to be responsible for assembling the segmented genome of influenza. In fact, influenza A virus genome is formed by eight different RNA segments and interestingly, each fully infectious viral particle contains precisely 8 RNA segments and 1 of each kind. By reducing the number of fully infectious viral particles, the severity of the disease and transmission is reduced.
Viral inclusions formed during IAV infection are particular, as these compartments do not exist in uninfected cells. In addition, viral inclusions display liquid properties, which means that the environment inside these compartments allows movement of the molecules. By concentrating progeny RNA in inclusions that are dynamic, each progeny RNA type can move to find its 7 additional partners, thus creating the right environment for its function.
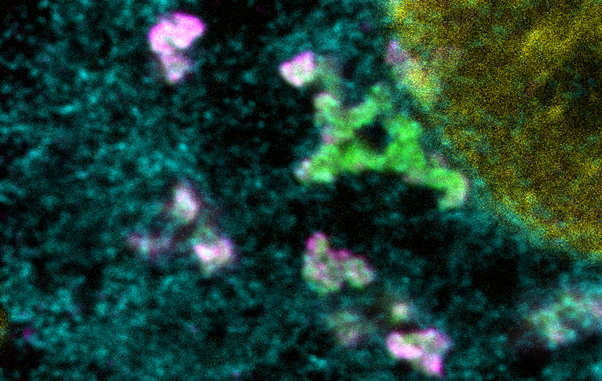
For the past 5 years, LOFlu has been studying how the host and the virus work in concert to coordinate the formation and material properties of IAV liquid inclusions and their interactions. This has revealed mechanisms of organisation of viral reactions in space and in time, ways to abrogate viral inclusion formation and to harden them. Our work has revealed that any perturbation on liquid inclusion formation or material properties reduces viral replication, making of them promising new antiviral targets against influenza A virus. This is especially interesting, as targeting them could be selective for infected cells without impacting healthy cells, having low toxicity. Thus, the project may provide new strategies to control season flu epidemics. Furthermore, given that many viral infections rely on such liquid material properties (e.g. measles, mumps, HIV, ebola) these findings may be translated to control other human relevant viruses.
Finally, the project has been identifying key principles governing the assembly of IAV pandemic genomes. Influenza A virus is a zoonotic virus and has a huge reservoir in aquatic and migratory birds. On occasion, avian influenza A virus can jump from birds to different species including to humans provoking pandemics. As the virus is very small, it is tightly adapted to one particular specie for efficient replication and transmission. Humans are different from birds, and these jumps require viral adaptations conferred by viral evolution. One mechanism to speed up this evolutionary process is conferred by mixing genomic parts of avian and human viruses in co-infections, in a process that involves the assembly process described above. Our project has been focusing on providing clues whether liquid viral inclusions serve to assemble mixed genomes. In addition, we are investigating whether viral inclusions are conserved amongst viruses infecting other species and revealing processes of viral adaptation critical for enabling host species jumps.
Collectively, LOFlu results will help control Influenza epidemics and pandemic outbreaks.
By Maria João Amorim, coordinatoor of LOflu and vice-director of Católica Biomedical Research Centre of Universidade Católica Portuguesa.
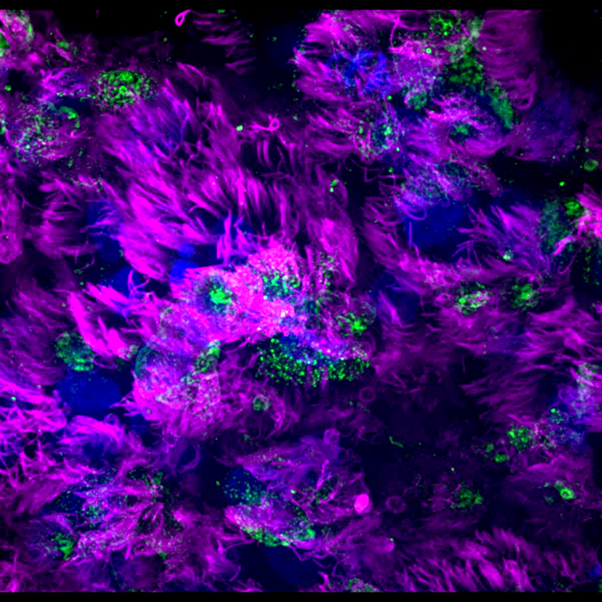
Image 1 - influenza A virus infected airway epithelial for 24h. Cilia in multiciliated cells are marked in magenta and the virus is marked in green, by marking a protein in influenza genome.
Image 2 - Hardened influenza A viral inclusions that lost their dynamic behaviour. The endoplasmic reticulum in cyan, viruses labeled in magente and Rab11a labelled in green.
Image 3 - Normal influenza A viral inclusions in infected cell that drive ithe assembly of segmented genome. The endoplasmic reticulum in cyan, viruses labeled in magente and Rab11a labelled in green.